The future of fighting infections
Researchers used the CLS to study proteins that a pathogen uses to break down sugar chains (glycans) present in human tissue during infections. This could lead to new treatments approaches for the bacterium.
By Wendy MatthewsDr. Boraston presenting at a symposium. Photo Credit: GlycoNet.
Scientists analyze 3D model of proteins from disease-causing bacteria at the CLS.
Millions of people are affected by the Streptococcus pneumoniae bacterium, which can cause sinus infections, middle ear infections and more serious life-threatening diseases, like pneumonia, bacteremia, and meningitis. Up to forty percent of the population are carriers of this bacterium.
Researchers from the University of Victoria used the Canadian Light Source at the University of Saskatchewan to study proteins that the pathogen uses to break down sugar chains (glycans) present in human tissue during infections. These proteins are key tools the bacterium uses to cause disease.
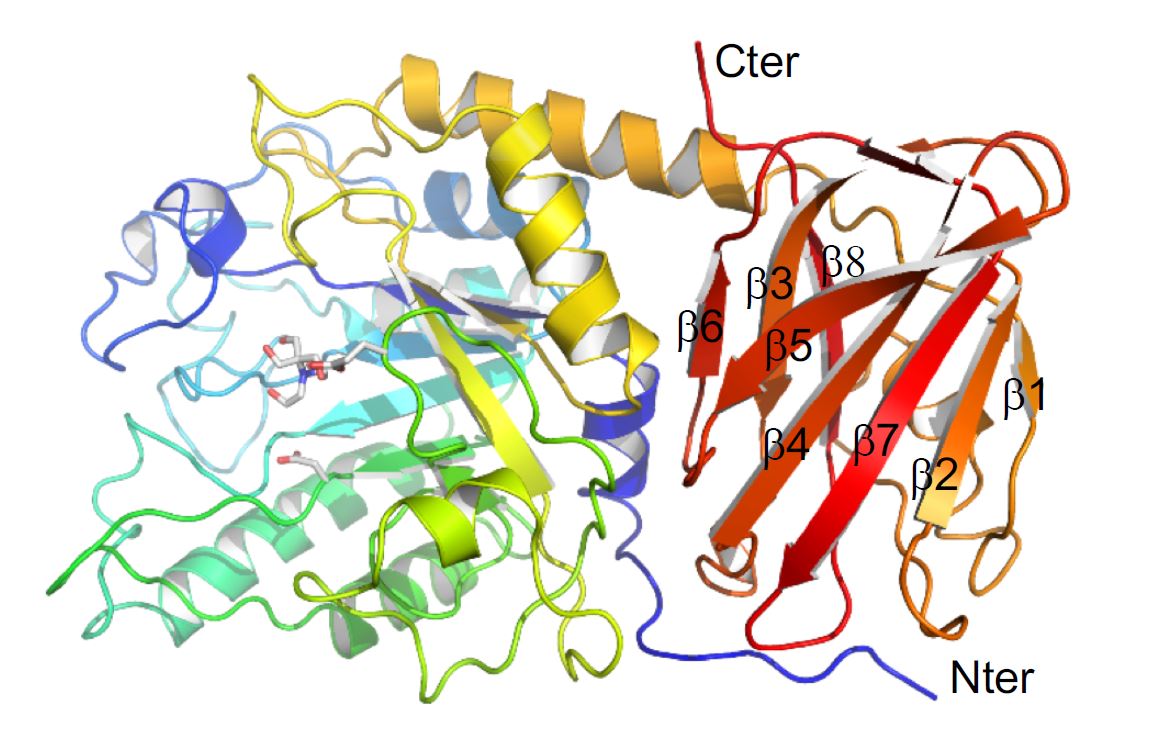
They used the Canadian Macromolecular Crystallography Facility (CMCF) at the CLS to determine the three-dimensional structure of a specific protein, an enzyme, that the bacterium produces to figure out how it interacts with and breaks down glycans.
Dr. Alisdair Boraston, one of the researchers from UVic, explained that the process is similar to a tree being chopped down. Where a complex glycan present in host tissue is the tree; the enzyme under study is like an axe that removes branches of the tree. In this case, it removes fucose from specific glycans, exposing the trunk of the tree (the core structure of the glycan), which then allows the trunk to be completely removed (by other enzymes).
This study, therefore, places this new enzyme into the larger context of enzymes that S. pneumoniae deploys during infection. Ultimately, his team wants to know if stopping this initiating enzyme would prevent the next enzyme from causing more damage.
“The question we were asking was partly if the enzyme is able to do this, but mostly how the enzyme acts,” says Dr. Boraston, which is why a 3D structural analysis of the protein using data collected at the CMCF was so important.
Now that they have analyzed this enzyme, it opens the pathway to developing new inhibitors and, eventually, possible therapies targeting the enzyme or other enzymes in the cascade may be developed.
“Our research is thinking ahead to alternative treatment approaches for a bacterium that is constantly adapting to medical interventions. We live in an era where the Streptococcus pneumoniae bacterium is becoming increasingly drug resistant. It is also continuing to change and evade our vaccination strategies. Thinking differently and considering alternative approaches to antibiotics is one possible future for fighting infections,” said Boraston.
By examining the pieces of this puzzle, and working to assemble them, the UVic research team are tackling one of the most prevalent worldwide threats to health.
This research was originally published in the Journal of Biological Chemistry. Hobbs, Joanne K., Benjamin Pluvinage, Melissa Robb, Steven P. Smith, and Alisdair B. Boraston. "Two complementary α-fucosidases from Streptococcus pneumoniae promote complete degradation of host-derived carbohydrate antigens." Journal of Biological Chemistry (2019): jbc-RA119. © the Author(s). DOI:https://doi.org/10.1074/jbc.RA119.009368
For more information, contact:
Victoria Schramm
Communications Coordinator
Canadian Light Source
306-657-3516
victoria.schramm@lightsource.ca
or
Jennifer Kwan
Senior Research Communications Officer
University of Victoria
Victoria, B.C.
250-721-7641
researchcomm@uvic.ca
