Scientists discover potential method to starve the bacteria that cause Tuberculosis
By deepening our understanding of how Tuberculosis bacteria feed themselves, University of Guelph researchers have identified a potential target for drug treatment.
By Greg BaskyA doctor uses a stethoscope on a child's back.

By deepening our understanding of how Tuberculosis bacteria feed themselves, University of Guelph researchers have identified a potential target for drug treatment. The team used the Canadian Light Source (CLS) at the University of Saskatchewan to image the bacteria in fine detail.
The infectious disease Tuberculosis (TB) is one of the leading causes of death worldwide. While rates of TB in Canada have remained relatively static since the 1980s, the disease disproportionately affects Indigenous populations. With TB-causing bacteria becoming increasingly resistant to antibiotics, researchers and drug makers are eager to find new, more effective treatments.
Researchers have known for some time that the bacteria that causes TB (Mycobacterium tuberculosis) uses our body’s cholesterol – a steroid – as a food source. Other relatives of the bacteria that do not cause disease share its ability to break down steroids. In this study, the University of Guelph team identified the structure of an enzyme (acyl CoA dehydrogenase) involved in steroid degradation in another member of the same bacteria family, called Thermomonospora curvata.
Dr. Stephen Seah, a member of the research team, said determining the structure of enzymes that metabolize steroids moves scientists and pharmaceutical companies one step closer to creating drugs that can inhibit a similar enzyme found in M. tuberculosis, which would effectively starve TB of its food source. The findings were recently published in the journal Biochemistry.
Knowing what an enzyme looks like – its structure – allows scientists to customize the shape of a drug to the enzyme target. Without the structure as a roadmap, scientists often end up exploring a lot of dead ends before arriving at a drug that fits its enzyme target tightly. Using the CMCF beamline at the CLS, the team was able to create a picture of the “keyhole” into which drug molecules need to fit.
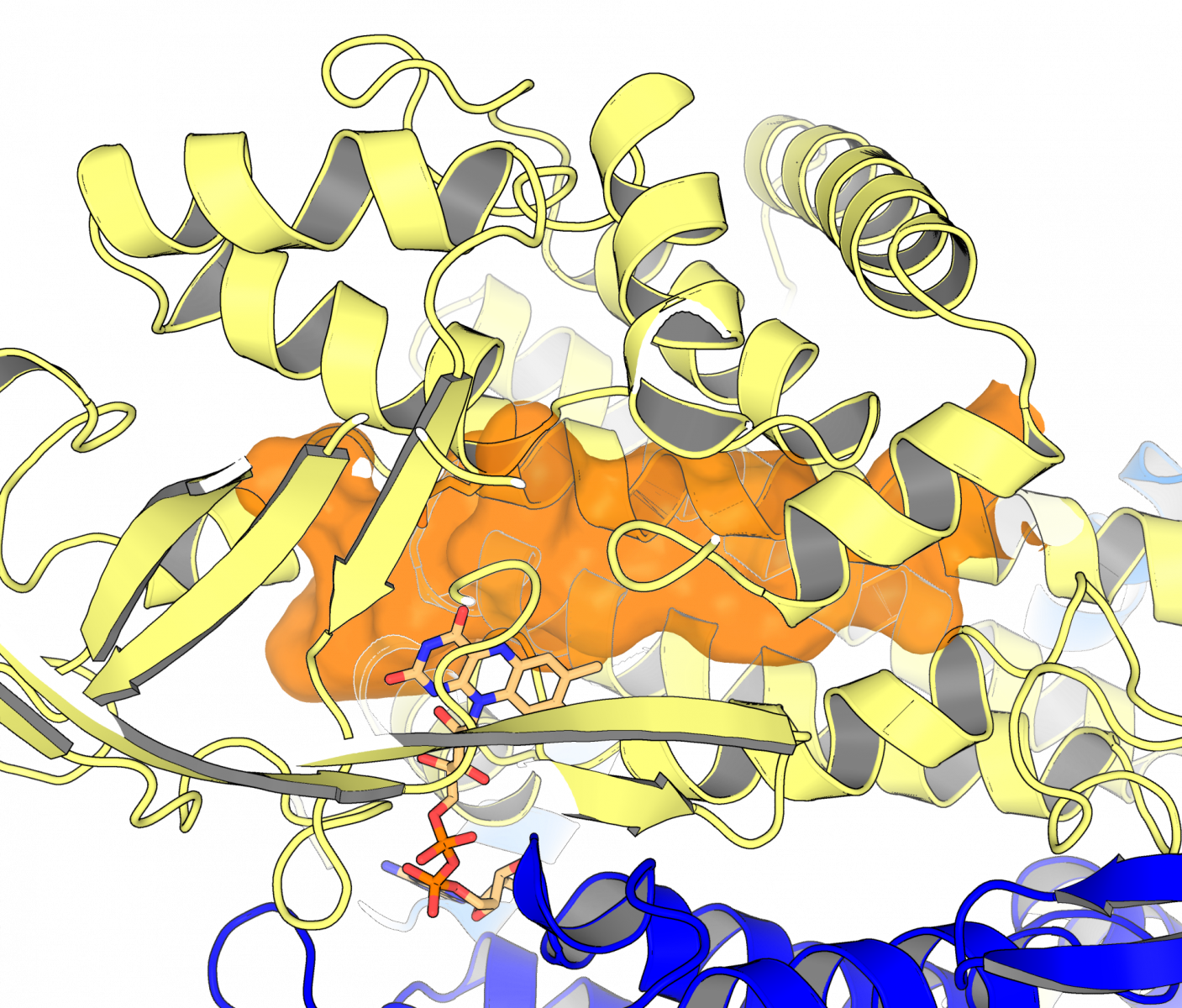
Dr. Matthew Kimber, another member of the University of Guelph team, said their findings help understand how this enzyme can be targeted. “We were surprised to observe that these enzymes are unusually adept at shifting their shapes as they go about their various tasks,” said Kimber. “This work helps us understand the exact shape of the keyhole a drug would need to fill to stop this enzyme in its tracks.”
Their discovery, said Seah, would not have been possible without access to the CLS beamline. “We depend heavily on the CLS for the X-ray source to determine the structure of our enzymes,” said Seah. “There’s a strong relationship between how bright the X-rays are and how much detail you can see in the final images.”
The team’s findings could also help drug makers create new steroid-related drugs such as anti-inflammatory or anticancer drugs. “This should help in building a toolbox for making new steroid drugs, or making the ones we do use more efficiently,” said Kimber.
Stirling, Alexander J., Stephanie E. Gilbert, Megan Conner, Evan Mallette, Matthew S. Kimber, and Stephen YK Seah. "A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity." Biochemistry (2020). DOI: 10.1021/acs.biochem.0c00568.
For more information, contact:
Victoria Schramm
Communications Coordinator
Canadian Light Source
306-657-3516
victoria.schramm@lightsource.ca
