Fighting antibiotic resistance
Researchers from the Universities of Guelph and Alberta used the CLS to identify a possible target in the prevention and treatment of bacterial infections.
By Erin MatthewsAntibiotic pills arranged into a map of the world.
The fight against infectious disease urgently requires new targets and treatments. The World Health Organization calls antibiotic resistance one of the biggest threats to global health, food security, and development.[1] It is crucial to find new ways of tackling infections and to gain a deeper understanding of bacteria themselves.
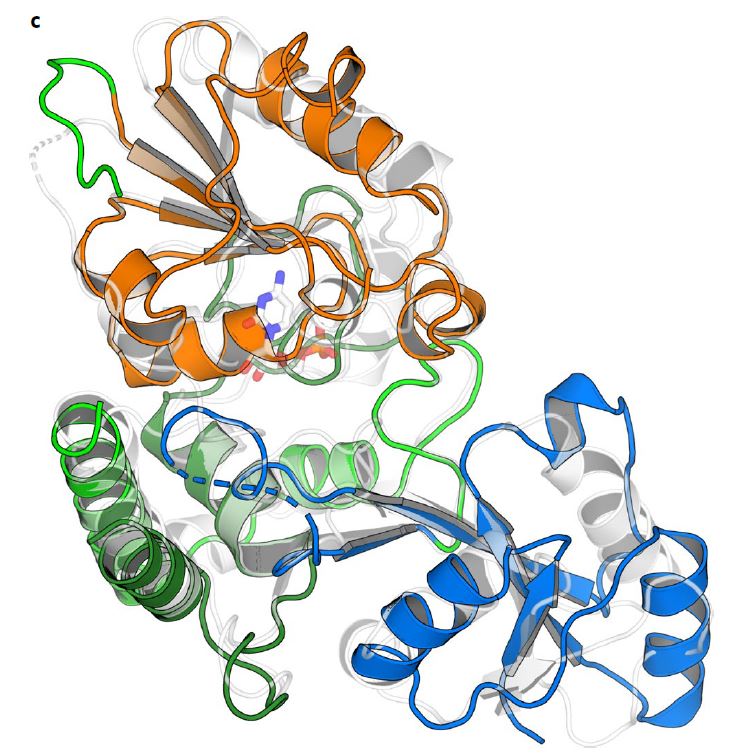
Chris Whitfield and Matthew Kimber, scientists from the University of Guelph, together with Todd Lowary from the University of Alberta have made great strides in understanding a critical component of bacterial capsules by using the Canadian Light Source (CLS) at the University of Saskatchewan. The capsules act as armour to shield the microbes from our immune system defenses.
While these capsules play a role in protecting pathogens like Neisseria meningitidis—the bacteria responsible for meningococcal disease—they are also a good target for therapeutics like small molecule inhibitors and vaccines.
If you get rid of the capsule, then the bacteria become very susceptible to attacks by the immune system, helping patients to get the upper hand during infections.
“The idea is that you identify molecules that inhibit one of the early enzymes. This prevents bacteria from making their capsule coats and renders them susceptible to normal host defenses,” Whitfield said.
“This is an approach called ‘anti-virulence’— the compounds don’t kill the bacteria, but they eliminate a key component required for survival and virulence in the host.”
The enzymes that Whitfield, Kimber and their team investigated are commonly found amongst a range of pathogens, making it a great target for different therapeutic techniques.
Whitfield’s group has been working to understand the details and mechanisms behind the assembly and export of these surface molecules for decades. While there have been clues in the scientific literature for years, more pieces of the puzzle are finally now coming together, offering a more detailed view of these processes.
The CMCF beamline at the CLS helped the researchers understand the structure of one of the enzymes, KpsC, through X-ray diffraction techniques.
“The CLS was critical for this work as all the structural data sets used were collected there,” Kimber said. “CLS is our preferred venue as it bypasses all of the uncertainty and complexity of shipping crystals internationally. It is very easy to use, is generally available on a reasonable time scale, has great support staff and has world-class technical capabilities.”
Isolating a target is a huge step towards the creation of clinical applications, but it is still a long journey to a tailored-fit therapy. Whitfield would like to get as far as possible so that pharmaceutical companies can be able to use this target in drug development.
He emphasized the importance of funders like the Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council, and Canadian Glycomics Network.
Both Kimber and Whitfield stressed the importance of understanding the intricacies of bacterial systems. The researchers hope to better understand how enzymes interact with the cellular machinery inside different bacteria species and to keep people healthier in the process.
Doyle, Liam, Olga G. Ovchinnikova, Katharine Myler, Evan Mallette, Bo-Shun Huang, Todd L. Lowary, Matthew S. Kimber, and Chris Whitfield. "Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules." Nature chemical biology 15, no. 6 (2019): 632. https://doi.org/10.1038/s41589-019-0276-8.
[1] https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance
For more information, contact:
Victoria Schramm
Communications Coordinator
Canadian Light Source
306-657-3516
victoria.schramm@lightsource.ca
